Date: 2019-09-26
Light is the most important energy source for plants and also modulates many developmental programs such as seed gemination, phototropism and flowering. Understanding how plants sense light to control growth and development can impact on crop production. A research team led by Dr. Shih-Long Tu at the Institute of Plant and Microbial Biology, Academia Sinica recently discovered a new mechanism to demonstrate how light regulates gene expression in plants. The breakthrough discovery may shed light on agricultural applications. Their results have been published in the scholarly journal “The Plant Cell” on Aug 13th.
To optimize light absorption, plants have evolved sophisticated photoreceptor systems to sense the quality, quantity, direction, and duration of light. Phytochromes are a major class of photoreceptors that mainly perceive red and far-red light. Phytochromes function at various levels to control gene expression such as during chromatin modification, transcription, translation, and post-translation. Accumulating evidence suggests that alternative splicing at post-transcriptional level is also regulated by phytochromes. However, the detailed mechanism remains unclear.
Alternative splicing is a widespread mechanism in eukaryotes in which two or more different forms of mRNAs are generated from a single gene. It adds a new level of complexity to the transcriptome and proteome. Alternative splicing has been extensively studied in animal system. It is known that most human genes are subject to alternative splicing. Many heritable diseases are caused by defects in alternative splicing. Although it is also abundant in plants and potentially involves in controlling developmental programs and environmental responses, detailed mechanism is still largely known.
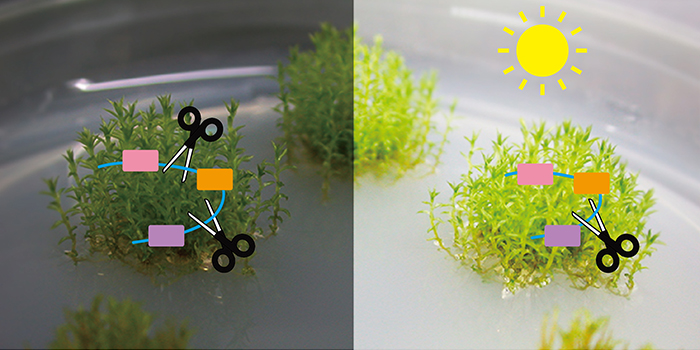
Dr. Tu’s team previously demonstrated that phytochromes participate in the regulation of alternative splicing in the model moss species Physcomitrella patens. In this study, they further provide molecular evidence that the moss phytochrome interacts with a splicing regulator named PphnRNP-H1 in the nucleus in a red light-dependent manner. PphnRNP-H1 also shows red light-stimulated, phytochrome-dependent binding with a spliceosomal component PpPRP39-1. Genome-wide analyses demonstrated the involvement of both PphnRNP-H1 and PpPRP39-1 in light-mediated splicing regulation. Their findings strengthen the hypothesis that phytochromes directly participate in the regulation of pre-mRNA splicing by controlling splicing activity. The interaction cascade from phytochrome to spliceosome provides a regulatory pathway by which photoreceptors control the activities of pre-mRNA splicing machinery and modulate plant growth and development.
This study was supported by Ministry of Science and Technology and Academia Sinica. The first author is Ms. Chueh-Ju Shih, a Ph. D. student in Molecular and Biological Agricultural Sciences Program, Taiwan International Graduate Program, Academia Sinica. Other authors include Hsiang-Wen Chen, Hsin-Yu Hsieh, Yung-Hua Lai, Fang-Yi Chiu, Yu-Rong Chen, and Shih-Long Tu. Detailed information about this publication can be found as below.
Title: Heterogeneous Nuclear Ribonucleoprotein H1 Coordinates with Phytochrome and the U1 snRNP Complex to Regulate Alternative Splicing in Physcomitrella patens Link: http://www.plantcell.org/content/early/2019/08/13/tpc.19.00314?rss=1
-
Ms. Pei-Chun Kuo, Media Team, Secretariat, Central Administrative Office, Academia Sinica
(02) 2789-8821,deartree@gate.sinica.edu.tw
-
Chang-Hung Chen, Public Affairs Section, Secretariat, Academia Sinica
(02) 2789-8059,changhung@as.edu.tw









 Home
Home