Date: 2025-11-10
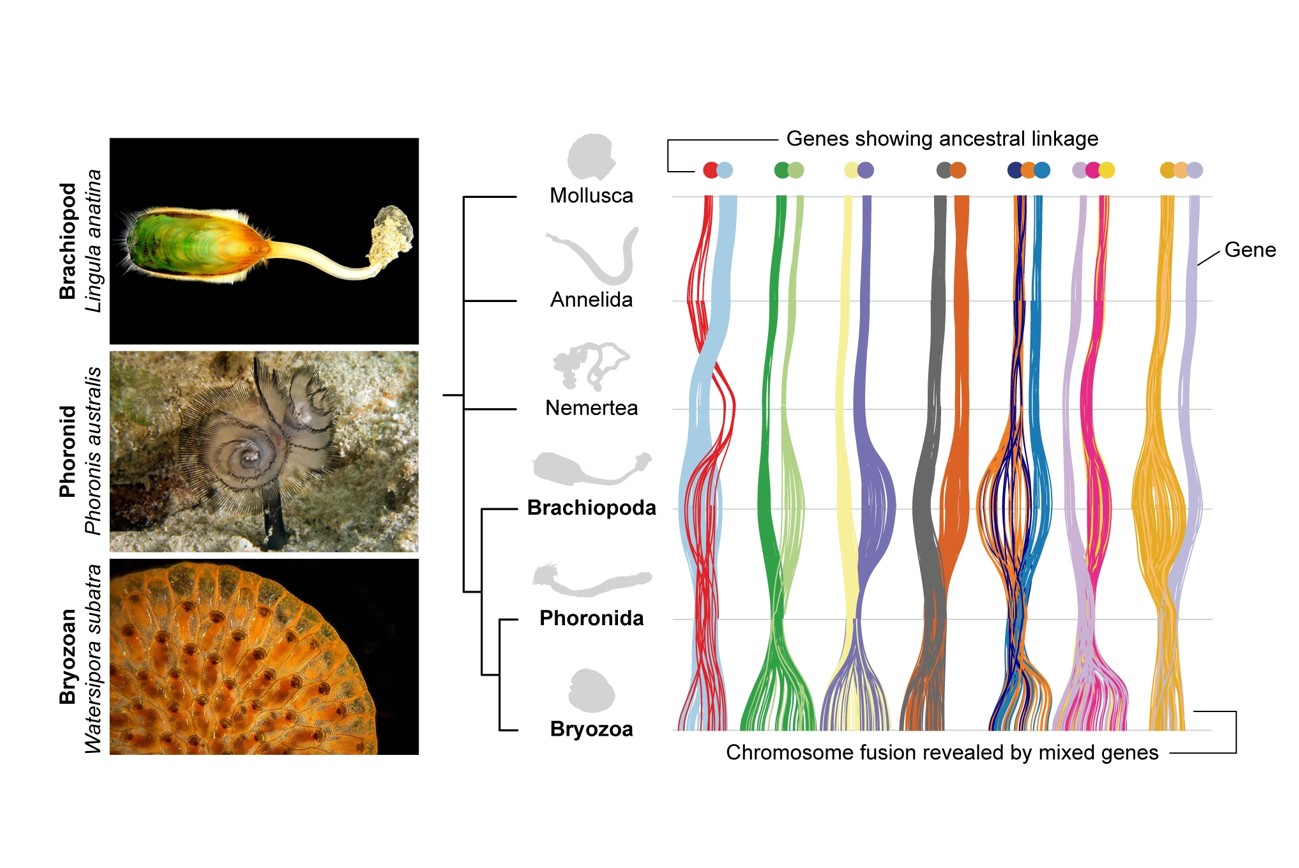
Resolving the deep branches of the animal tree of life has long been a major challenge in evolutionary biology. One of the most enduring debates concerns the classification of Lophophorata, a group that includes brachiopods, phoronids, and bryozoans, all characterized by a ciliated feeding structure known as the lophophore. While some genomic studies have supported their monophyly, others have suggested that bryozoans should be grouped with different lineages, and the controversy has persisted for more than a century.
A team led by Assistant Research Fellow Yi-Jyun Luo at the Biodiversity Research Center, in collaboration with researchers in Japan and the United Kingdom, has now generated the first chromosome-level genome of a phoronid and compared it with those of bryozoans and brachiopods. The study uncovered seven irreversible chromosome fusion events shared between phoronids and bryozoans, one of which is also shared with brachiopods. These fusions are absent in other lophotrochozoans such as molluscs, annelids, and nemerteans, providing sequence-independent evidence for the monophyly of lophophorates and demonstrating that these three phyla indeed share a recent common ancestor.
In addition, phylogenomic analyses and transcriptome sequencing of lophophores revealed that the three groups not only share a common set of genes, but also co-opt the same ancestral program involving vertebrate head-development genes. This shows that lophophores are homologous organs rather than convergent filter-feeding adaptations. This study resolves the century-old debate over the monophyly of lophophorates and highlights the power of genome structure analysis in reconstructing deep evolutionary relationships. The findings were published in Current Biology in November 2025 and featured as the cover story. The study was supported by the Academia Sinica Career Development Award and the National Science and Technology Council Research Project Grant.
-
Link









 Home
Home